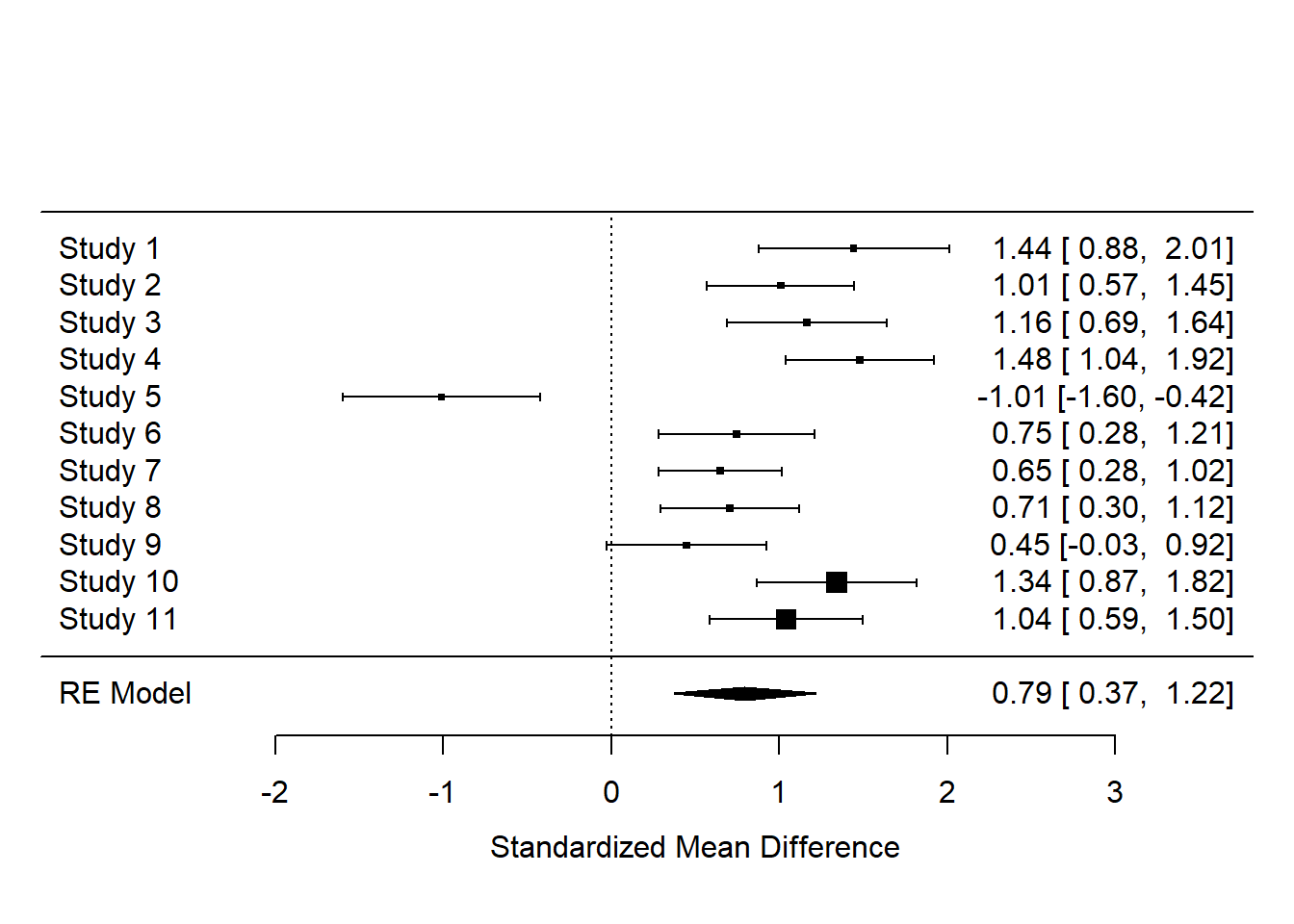
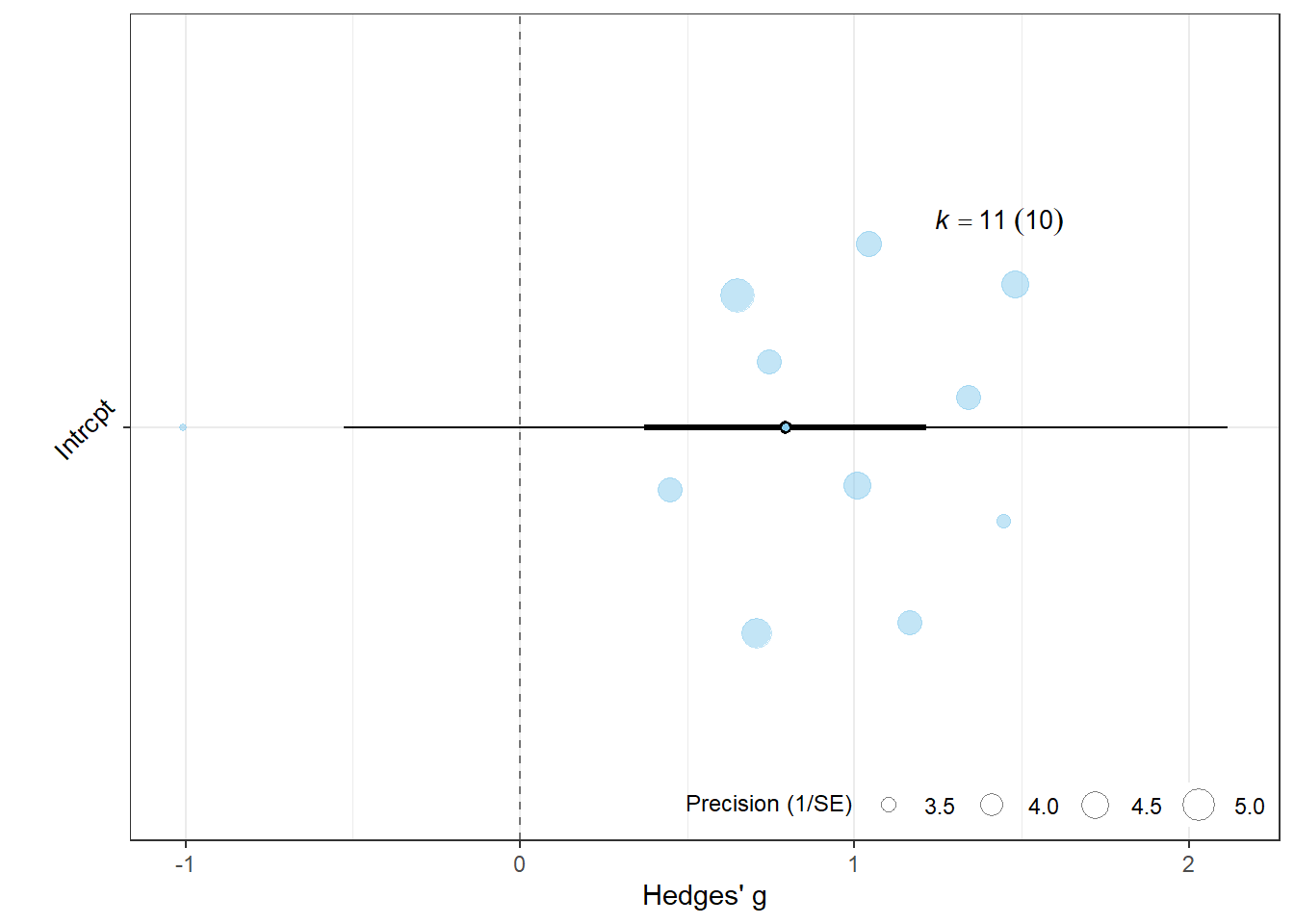
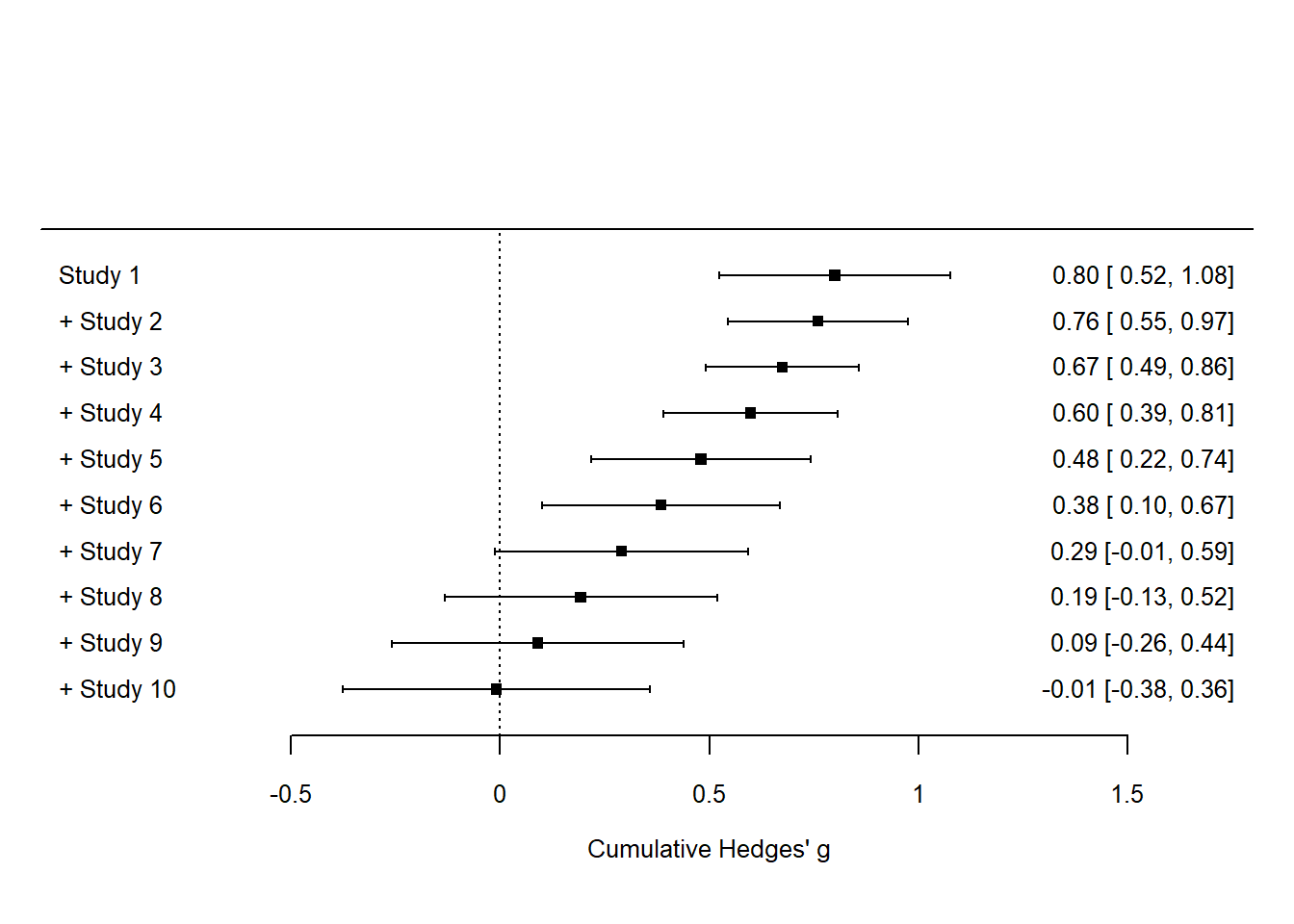
data <- data.frame(
study_id = c(1:10,10),
# Mean species richness in restored and control sites
mean_restored = c(12, 15, 14, 20, 8, 13, 16, 19, 10, 18, 17),
mean_control = c(9, 12, 11, 15, 10, 11, 14, 17, 9, 14, 14),
# Standard deviations for restored and control groups
sd_restored = c(2.1, 3.0, 2.5, 3.5, 2.0, 2.8, 3.2, 2.9, 2.3, 3.1, 3),
sd_control = c(2.0, 2.9, 2.6, 3.2, 1.9, 2.5, 2.9, 2.7, 2.1, 2.8, 2.7),
# Sample sizes for restored and control groups
n_restored = c(30, 45, 40, 50, 25, 38, 60, 48, 35, 42, 42),
n_control = c(30, 45, 40, 50, 25, 38, 60, 48, 35, 42, 42)
)
data |> head() study_id mean_restored mean_control sd_restored sd_control n_restored
1 1 12 9 2.1 2.0 30
2 2 15 12 3.0 2.9 45
3 3 14 11 2.5 2.6 40
4 4 20 15 3.5 3.2 50
5 5 8 10 2.0 1.9 25
6 6 13 11 2.8 2.5 38
n_control
1 30
2 45
3 40
4 50
5 25
6 38